Sponsored by Lantern Pharma
Powering Cancer Drug Discovery and Development Using the Artificial Intelligence and Machine Learning Platform RADR®
Introduction
Glioblastomas (GBM) are devastating brain tumors and rank as one of the most aggressive human cancers with a median overall survival of 15 months. In the U.S., there are around 15,000 patients diagnosed with GBM annually. Unfortunately, over the last two decades there have been no effective therapies developed to treat GBM outside of the standard-of-care (SOC) agent temozolomide (TMZ). The effectiveness of TMZ is limited to only around 30-50% of patients who have tumors which have a methylated promoter of the DNA repair enzyme known as O6 Methyl Guanine Methyl Transferase (MGMT), leaving around 50-70% of patients without available therapy. GBM patients that have unmethylated status or relapse from TMZ therapy have no effective treatment options available and have very poor outcomes.
One of the limitations to the discovery of new agents to treat GBM is the ineffectiveness, high costs, and lengthy timelines it takes for traditional oncology drug discovery and development. The average time to bring a drug into clinical trials can take between 3-7 years, $30-50 million, and can have a failure rate of up to 96.7% in clinical trials, all which limit the progression of bringing novel therapies to patients. Lantern Pharma is one of a handful of companies that is using artificial intelligence (A.I.), machine learning (M.L.), and large scale multi-omic data to shift the narrative of oncology drug discovery and development and rapidly overcome these hurdles.
Lantern’s proprietary RADR® A.I. and M.L. platform is one of the world’s largest oncology drug discovery and development platforms, consisting of over 25+ billion oncology focused data points. These data points consist of large scale transcriptomic and genetic data, over 130,000 patient records, 150+ drug-tumor interactions, and cover over 135 cancer types/subtypes. RADR® leverages this data and over 200+ advanced M.L. algorithms to power its 4 drug discovery modules which can 1) derive M.L. based drug response signatures, 2) discover a drug's mechanism(s) of action, 3) identify new indications for a drug and 4) determine optimal drug combinations (Fig. 1). Combined, RADR®’s data, capabilities, and insights are powering the development of Lantern’s drug programs from initial RADR® insight towards first in human clinical trials in around 2 years at markedly reduced costs. This has been exemplified in Lantern’s GBM program for its drug candidate LP-184.
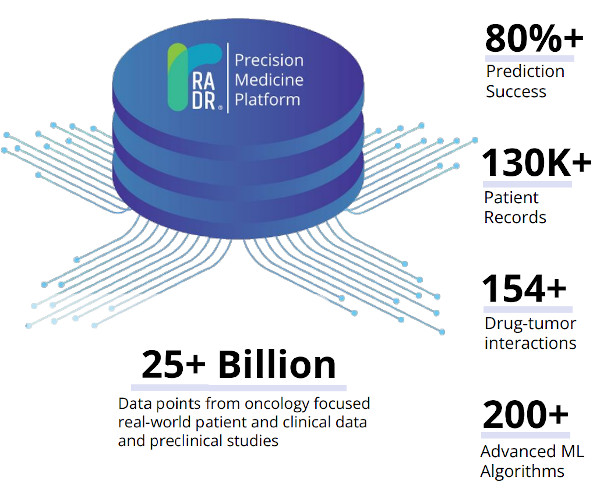
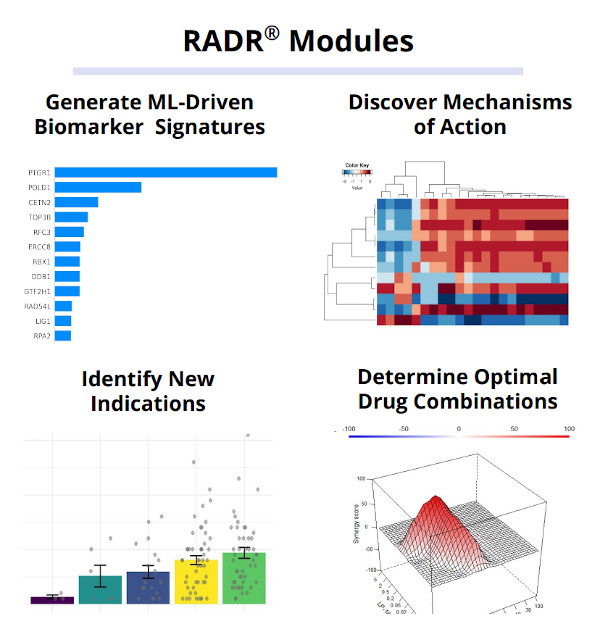
Figure 1. RADR® is an advanced A.I. and M.L. oncology focused drug discovery and development platform. It contains over 25 billion cancer focus datapoints that are utilized by RADR®’s four M.L. modules.
RADR® Insights Facilitate Development of Lantern’s Drug Candidate LP-184
LP-184 is a new generation of acylfulvene and DNA damaging agent that is a leading drug candidate in Lantern’s portfolio and is being developed for multiple solid tumors and central nervous system cancers. A key to the development of LP-184 has been RADR® M.L.-driven modeling and insights that have aided in discovering key aspects of LP-184’s mechanism of action and predicting cancers that are sensitive to LP-184. Highlighted here are several examples of the insights generated by RADR® that have accelerated the development of LP-184 for GBM.
RADR® M.L. Modeling Aides in Identifying PTGR1 as a Predictive Biomarker for LP-184
To generate a M.L. model and predictive biomarkers for LP-184, Lantern first generated new drug sensitivity data of LP-184 from the National Cancer Institute’s cancer cell line panel (NCI-60) cell lines, the Broad Institute’s Cancer Cell Line Encyclopedia (CCLE) cell lines, and patient derived xenograft (PDX) models. Large scale multi-omics data from these experiments was used in the automated RADR® M.L. pipeline to test multiple feature selection and model building algorithms to find the best predictive biomarkers and M.L. model for drug response prediction for LP-184.
The RADR® drug response signatures discovery M.L. pipeline builds thousands of models with millions of cross validation fits using hyper-parameter tuning. Hyper-parameter tuning allows for optimization to identify the best parameter combinations to tune the model, while cross validation allows for optimization to assess the effectiveness of the model and to reduce the overfitting. This automated pipeline has the capability to build thousands of response prediction models in less than a day and rank them based on their performance.
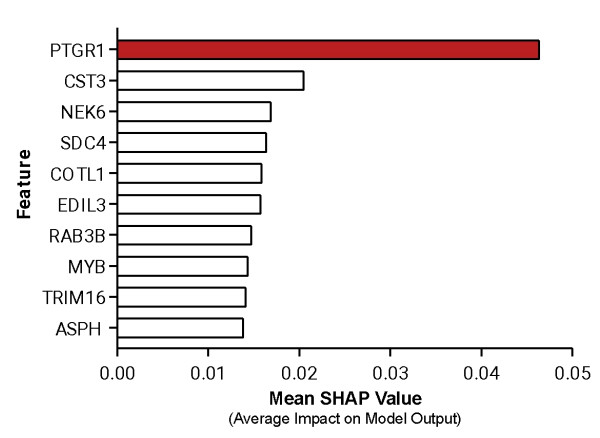
Figure 2. Top ten features from the RADR® M.L. model used to predict LP-184’s drug sensitivity, PTGR1 was the top-ranking feature driving the model’s prediction.
For LP-184, the M.L. model was trained on 77 samples with drug sensitivity and expression data, while keeping 33 samples (composed of a mixture of cell lines and PDX samples) completely blind for testing. As a result, the biomarker discovery M.L. pipeline selected the best performing model, with PTGR1 as the top ranking feature that drives the model prediction (Fig. 2). The best model was able to predict the drug sensitivity closer to the actual values with significant (p value = 8.48e-08) correlation coefficient of r = 0.78 (Fig. 3). The blind testing data consisted of 26 cell lines and 7 PDX samples. This trained and tuned M.L. model can predict LP-184 sensitivity on cancer cell lines, PDX models, and patient data.
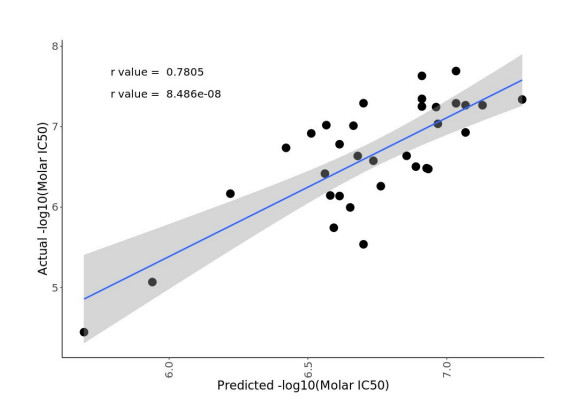
Figure 3. Model performance using blind testing data from 33 samples. The model’s predictions were significantly correlated with actual drug sensitivity values (p = 8.48e-08, r = 0.78).
RADR® M.L. Model Predicts GBM Tumors will be Sensitive to LP-184
Following the finding that the top gene signature corresponding to LP-184 sensitivity was PTGR1, additional bioinformatic analysis was performed by RADR® to predict what cancer types would be most sensitive to LP-184 treatment. Using patient gene expression data from The Cancer Genome Atlas (TCGA), RADR® predicted that PTGR1 overexpression in GBM and other high and low grade gliomas was higher than the RADR® estimated minimum PTGR1 expression level required for LP-184 sensitivity (Fig. 4), meaning that GBM and other high and low grade gliomas are likely to be sensitive to LP-184 treatment.
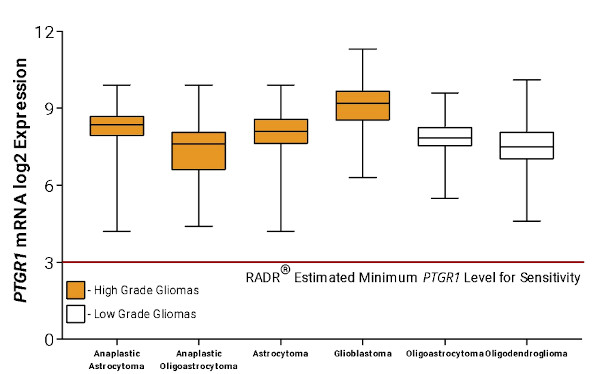
Figure 4. RADR® predicted that GBM, high-grade gliomas, and low-grade gliomas have sufficient PTGR1 expression to be sensitive to LP-184
The oncogene EGFR is involved in GBM tumorigenesis and malignancy, and 57% of primary GBM patients are reported to have gain-of-function alterations. Our preclinical data showed EGFR activity enhances likelihood of LP-184 response (table), which is consistent with our predictions of LP-184 response that showed a positive association of Normalized Enrichment Score (NES) of EGFR activity and predicted response. Nearly all GBM patients in the US are treated with temozolomide (TMZ), which favors tumor re-expression of MGMT through promoter demethylation and resistance to TMZ-induced DNA damage. TMZ methylation is used as a clinical marker for TMZ susceptibility, while TMZ demethylation and expression is a marker for TMZ resistance. We found TMZ methylation is negatively correlated to LP-184 response in cell lines. These data suggest LP-184 will have a promising target profile for the majority of GBM. The main obstacle to therapeutic efficacy and long term survival with GBM treatment is development of TMZ resistance through MGMT re-expression. As MGMT does not show a similar anti-correlation with LP-184, it suggests LP-184 can be a promising agent for treatment of TMZ-resistant GBM.
Validation of RADR® Insights in the Wet Lab
Following the RADR® insights that strongly predicted that LP-184 would be effective for GBM, Lantern conducted in vitro and in vivo experiments to validate RADR®’s predictions. LP-184 was found to have nanomolar potency across several genetically diverse GBM models, including both MGMT methylated and unmethylated GBM models. Additionally, LP-184 was found to have high blood brain barrier permeability similar to TMZ and have higher bioavailability in brain tumors compared to the surrounding brain tissue (2:1 ratio). These and subsequent preclinical experiments validated LP-184’s sensitivity for GBM and supported its progression towards a first in human Phase 1 clinical trial.
Conclusion
Implementing RADR®’s A.I. and M.L. insights into Lantern’s drug discovery and development platform has rapidly accelerated its pipeline of drug programs, including LP-184 for GBM. In around two years, Lantern has progressed its GBM program from initial RADR® insights, to wet lab validation, to late stage IND studies - significantly cutting typical drug development timelines and costs. Using this model of drug development, Lantern has achieved similar timelines for several of its other programs and is anticipating a Phase 1 clinical trial for GBM in early 2023.
Since the initial RADR® insights in 2020, RADR® has undergone significant growth across the entire platform, including the development of more sophisticated M.L. models, the incorporation of advanced ensemble algorithms, and significant upgrades to infrastructure. These advancements have increased the predictive accuracy and functionality of RADR® and continue to power the drug discovery development of Lantern and its collaborators.