Sponsored by Kymera Therapeutics
Identifying High Impact Tissue Sparing E3 Ligases and Their Binders
There are many drug target classes for which unwanted on-target pharmacology limits clinical application. Understanding E3 tissue expression across both healthy and diseased tissues enables a disease-directed TPD strategy based on all mapped E3 ligases. Using comparative analyses of expression patterns, one can identify selective pairings of E3 ligases with therapeutic targets of interest. Ligandability, Expression profile and Degradative activity (LED) criteria can be used to identify novel E3 ligases to match with clinically validated targets.
Fragment, structural, and virtual screening approaches have been used to identify ligands to a novel cullin E3 ligase, which were successfully made into degraders. The co-crystal structure of an initial fragment hit with a Kd of > 1 mM was solved by X-ray crystallography revealing interactions within the degron binding pocket of this ligase. SBDD allowed improvement in affinity to approximately 30 µM and further cycles of in silico library design and synthesis further improved the potency to nM hits with good physical properties and low molecular weight. Degraders of multiple proteins including BRD4 and STAT3 were discovered using this novel E3 ligase binder.
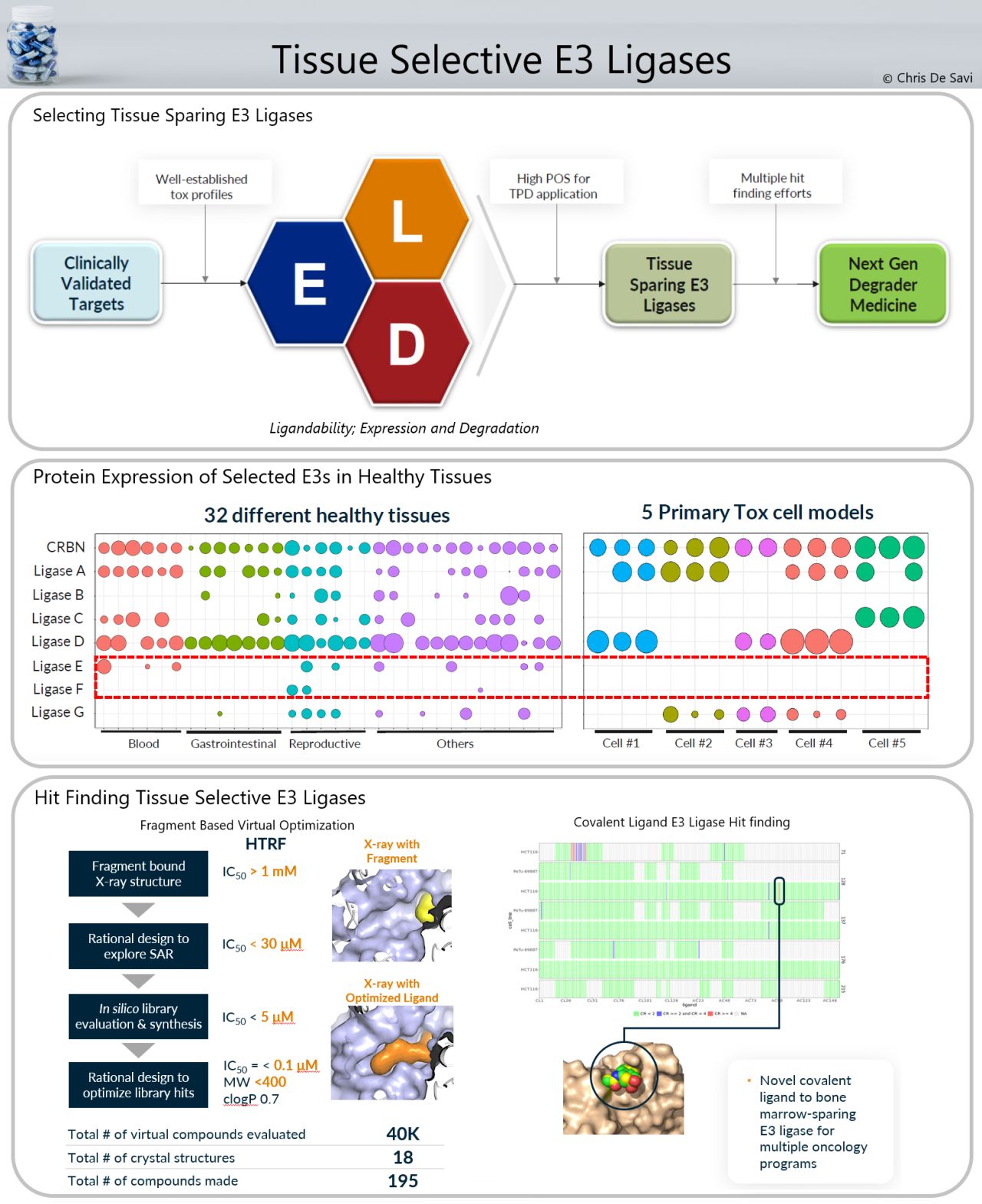
Figure 1: Fragment, structural, and virtual screening approaches.
Activity-based protein profiling (ABPP) to screen covalent compound libraries using chemoproteomics is another approach to find novel hits for E3s across the proteome within a single set of experiments. In one example, an X-ray structure of a small novel covalent hit bound to a specific cysteine residue of a tissue sparing E3 ligase is highlighted. These binders are being incorporated in multiple oncology targeted protein degraders where sparing bone marrow toxicity will be important for both efficacy and safety of future therapeutics.
The platform, developed by Kymera Therapeutics, can identify high impact tissue sparing E3 ligases that are lowly expressed or absent in target cells responsible for on-target toxicity, and to identify novel binders to these ligases. These binders will be incorporated into new targeted protein degrader medicines which will enable us to unlock clinically validated targets where on-target unwanted pharmacology limits the clinical utility of small molecule inhibitors.