Sponsored by Cyclica
Cyclica's Chemical Probes for DCAF1
Cyclica is designing chemical probes for DCAF1, a potentially novel target for protein degradation, with proprietary technology MatchMaker
DCAF1 was originally identified as a putative antiviral host target and more broadly as a promising target to facilitate proteasome-mediated degradation of therapeutic targets as a part of a E3 ubiquitin ligase complex. It has a complex domain architecture that includes a WD40 repeat (WDR) domain, which is one of the most abundant protein-protein interactions (PPIs) domains in the human proteome. Given the significant role that PPIs play in many cellular processes and diseases there has been renewed interest in exploring WDR domain proteins, including DCAF1.
Canadian data-driven biotech company Cyclica Inc. developed an AI-enabled deep learning platform capable of assessing small molecule-protein interactions across the proteome. DCAF1 recruits substrate proteins through its WDR domain to the CUL4A-RBX1-DDB1-DCAF1 E3 ubiquitin ligase complex for subsequent proteasomal degradation, which is a key process that will be scrutinized by small molecule probes. MatchMaker predicted small molecules that not only would bind DCAF1, but were also not likely to interact with undesirable off-targets. Using MatchMaker, Cyclica predicted and tested experimentally 100 compounds for DCAF1 in collaboration with the Structural Genomics Consortium (SGC). Compounds were assessed through Surface Plasmon Resonance to determine binding affinity to DCAF1, and a top compound, CYCA-117-70 showed selective binding for DCAF1. Subsequently, DCAF1 was co-crystallized with CYCA-117-70 and deposited as the first ligand-bound structure of DCAF1 in the PDB.
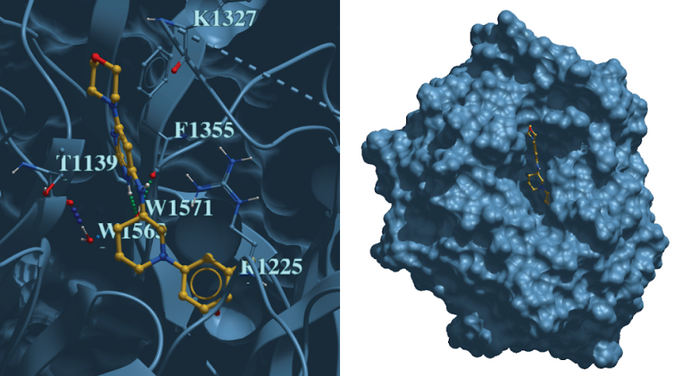
Figure 1: Co-crystal structure of CYCA-117-70 bound to the WDR-domain of DCAF1
With help of deep learning, Cyclica demonstrated a unique ability to discover binders for low data/undrugged targets — defined as proteins with very little structural information (or low resolution) and little to no chemical data (no co-crystal structure, no known bound ligands, etc). Unlike other approaches that require high quality structure with defined pockets, a co-crystal structure and ideally a large amount of chemical equity, Cyclica’s MatchMaker approach generalizes protein binding even to low data targets, opening the door for discovering drugs for highly unmet medical need.