Insilico Medicine's AI Designs ENPP1 Inhibitor for Immune Modulation in Cancer
A study published in Nature Communications reports the development of ISM5939, an orally active small-molecule ENPP1 inhibitor designed using Insilico Medicine’s generative AI platform.
The compound works by inhibiting ENPP1, an enzyme that suppresses immune activity in tumors, to reactivate the STING (Stimulator of Interferon Genes) pathway. According to the authors, this may help restore local immune responses in cancers that don’t respond to checkpoint inhibitors or chemotherapy.
Therapeutic Rationale: ENPP1 as an Immune Modulation Target
By blocking ENPP1, ISM5939 preserves cGAMP levels and reactivates the STING pathway in tumors, helping the immune system detect and fight cancer. Unlike direct STING agonists, which require intratumoral delivery and often trigger systemic inflammation, ISM5939 is designed to work orally with a better safety profile.
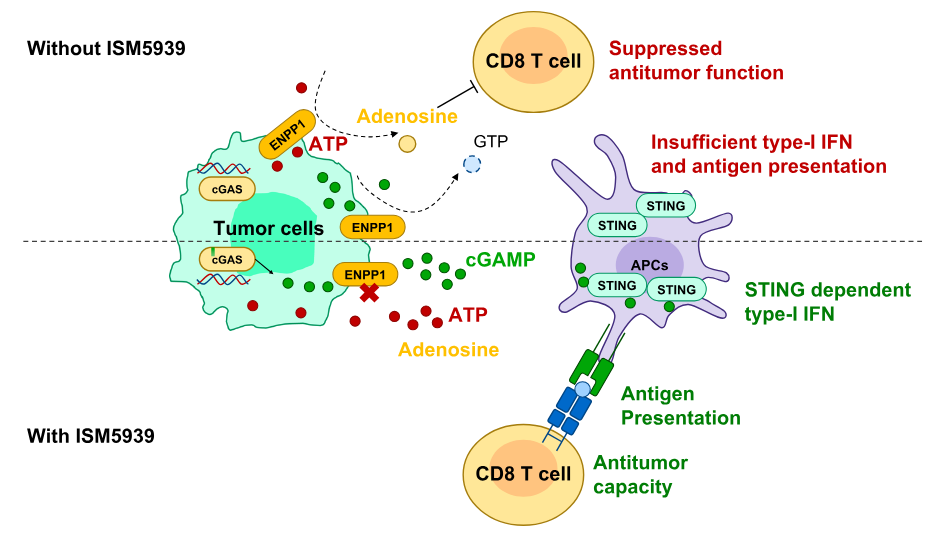
Mechanistic model of ISM5939: ENPP1 inhibition preserves extracellular cGAMP and reduces adenosine levels, activating STING signaling in APCs and enhancing CD8+ T cell responses. See study for details.
The study further reports that for the first time an ENPP1 inhibitor was able to overcome dual resistance to both immune-checkpoint blockade and chemotherapy in preclinical models.
Computational Design and Optimization Process
Insilico used its PandaOmics platform to prioritize ENPP1 as a target based on transcriptomic data from TCGA and spatial/single-cell analyses. Chemistry42, the company's structure-based generative design engine, generated candidate molecules from scratch, with initial hit compounds identified within three months of project initiation.
Alchemistry modules were applied to predict binding affinity, while additional models screened for ADMET properties, hERG risk, and on-target potency.
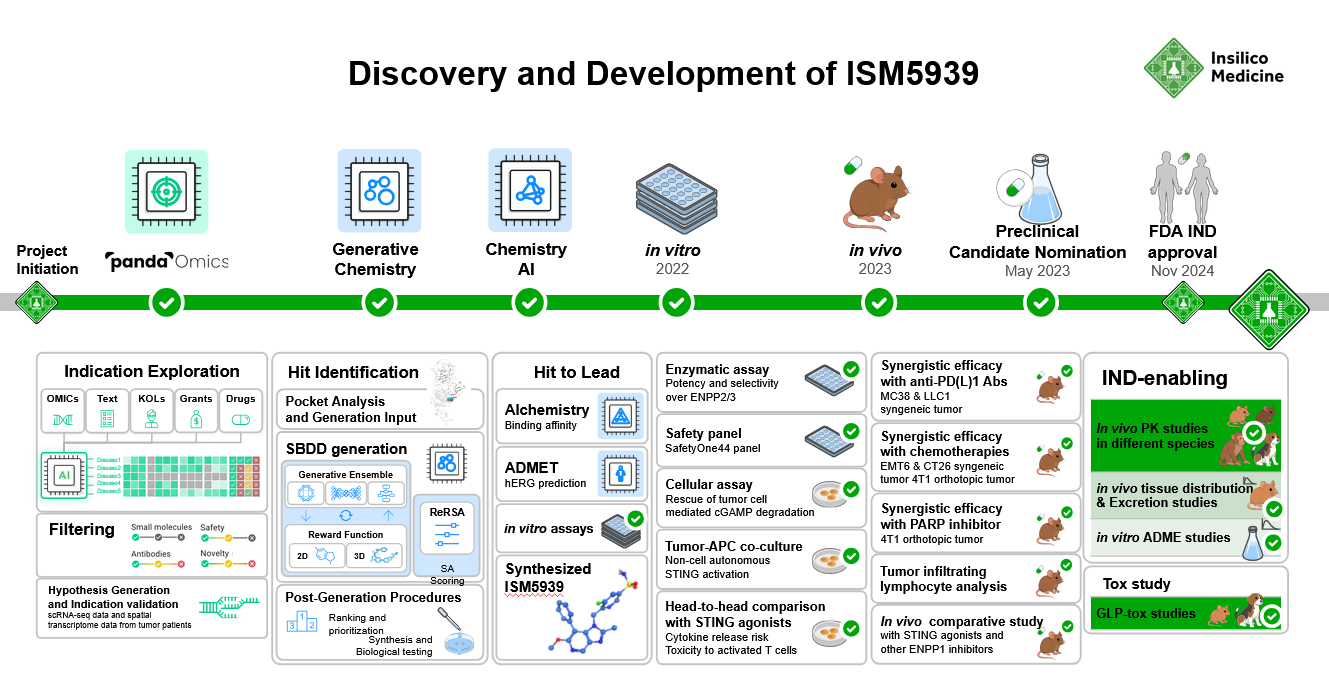
Starting from known ENPP1 scaffolds, the team generated hit compounds and refined them over several iterations. ISM5939 was selected based on predicted drug-like properties and in vitro potency.
Preclinical Findings
In preclinical tumor models, ISM5939 enhanced the efficacy of checkpoint inhibitors (e.g., anti-PD-1), DNA-damaging chemotherapies, and PARP inhibitors. It increased cGAMP accumulation in tumors, promoted STING activation in immune cells, and avoided systemic pro-inflammatory cytokine induction. The compound was also reported not to induce effector T cell death in the tumor microenvironment.
ENPP1 expression was found to be high in triple-negative breast cancer, hepatocellular carcinoma, colorectal adenocarcinoma, acute myeloid leukemia, ovarian cancer, ER-negative breast cancer, breast cancer, and head and neck cancer. The study suggests these indications may benefit from targeted ENPP1 inhibition, either as monotherapy or in combination with existing immune-oncology agents.
The authors note that while immune checkpoint inhibitors have reshaped oncology, only approximately 10–35% of treated patients achieve durable clinical responses.
Context
The study is the third Insilico publication in a Nature Portfolio journal in 2025, and the fifth since 2024 focused on AI-designed drug candidates. Earlier work includes Rentosertib (TNIK inhibitor for fibrosis) and ISM5411 (PHD1/2 inhibitor for IBD), both published in Nature Biotechnology. In January 2025, Insilico and the University of Toronto jointly reported a method for KRAS inhibitor design using a quantum-classical hybrid model.
See also: Insilico Medicine Reports Benchmarks for its AI-Designed Therapeutics
Between 2021 and 2024, Insilico nominated 22 preclinical drug candidates. According to the company, each program required the synthesis of approximately 60 to 200 compounds and reached nomination within 12 to 18 months. This compares to the 2.5 to 4 years typically required in conventional small-molecule discovery. Insilico states that all nominated candidates advanced to the IND-enabling stage.
Topics: AI & Digital