Owkin Shares Federated Learning Tool for Secure RNA-Seq Analysis
Owkin has released FedPyDESeq2, a federated learning tool for differential expression analysis (DEA) in bulk RNA sequencing (RNA-seq). Designed to address challenges in large-scale transcriptomics studies, such as data privacy and siloed datasets, the tool enables collaborative analysis without sharing sensitive data. The tool is open-source, providing researchers with a practical solution for secure and effective DEA.
A New Approach to RNA-Seq Analysis
RNA-seq studies hold significant potential for clinical research, but progress is often limited by two key factors:
- Data Silos: Strict privacy regulations prevent the pooling of data from multiple institutions, reducing statistical power.
- Privacy Risks: Protecting sensitive genomic data remains a critical challenge in collaborative research.
FedPyDESeq2 addresses these issues by applying federated learning, which allows institutions to perform joint analyses while keeping all raw data and sensitive information secure on-site.
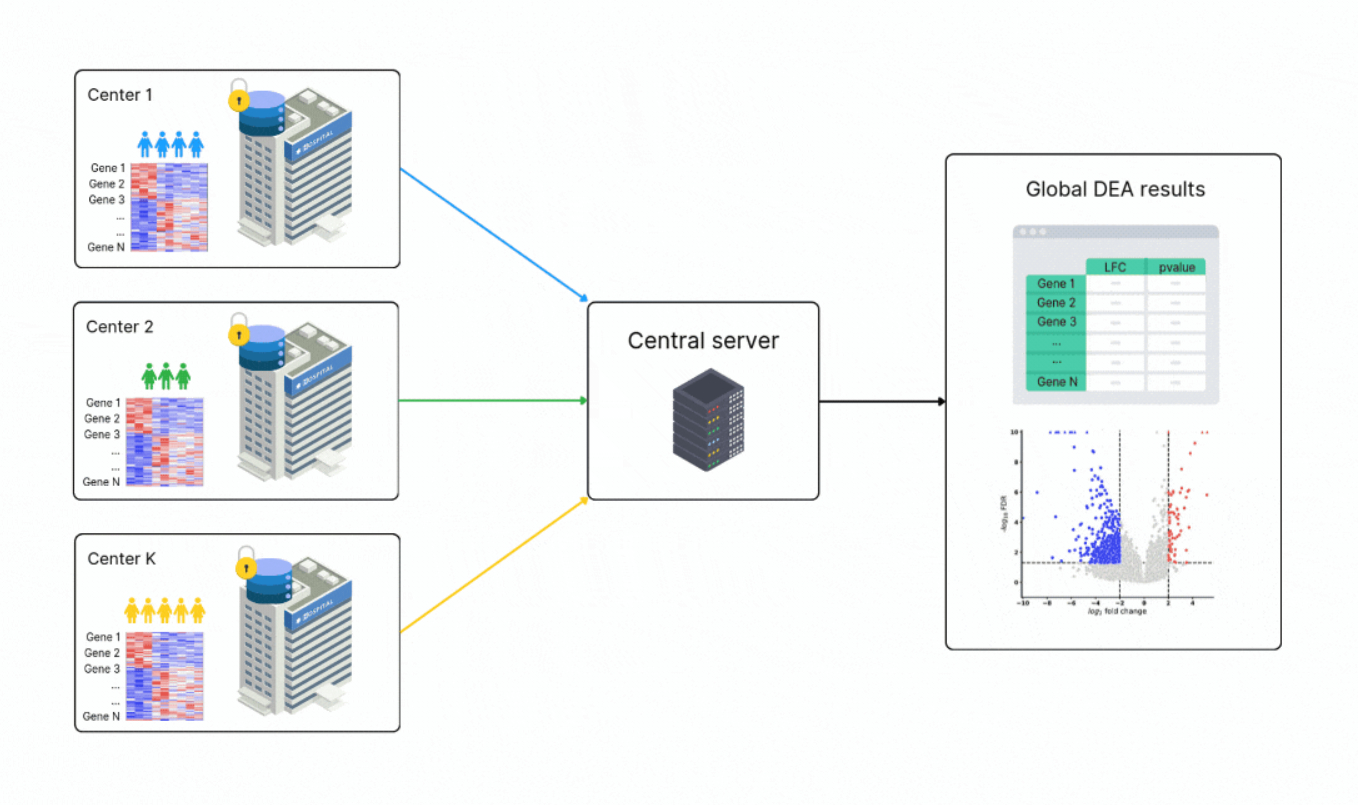
Federated workflow for differential expression analysis; Credit: Owkin
Built on PyDESeq2, a Python-based reimplementation of the widely used DESeq2 methodology by Love, Huber, and Anders (2014), FedPyDESeq2 ensures consistency with established DEA standards while extending its usability to distributed data environments.
Key Features
- Performance: FedPyDESeq2 produces results closely aligned with pooled PyDESeq2 analyses and outperforms traditional meta-analysis methods in terms of accuracy and sensitivity.
- Flexible Design: Supports multi-factor experimental designs, categorical and continuous covariates, and incorporates advanced outlier detection methods.
- Open-Source: Available on GitHub, along with scripts for reproducing experiments and benchmarks.
- Compatibility: Built using the federated learning platform Substra, FedPyDESeq2 replicates the steps of the DESeq2 pipeline in a distributed setting, ensuring minimal deviation from pooled data workflows.
Why It Matters
Owkin’s FedPyDESeq2 provides a way to conduct DEA on siloed datasets, enabling collaborative research across institutions while maintaining compliance with privacy regulations. This tool opens possibilities for:
- Cohort Comparisons: Performing DEA across fully partitioned datasets, such as comparing patient groups from different hospitals.
- Improved Data Security: Avoiding raw data exchanges while achieving near-identical results to pooled analysis.
At the same time, the authors acknowledge potential challenges: while FedPyDESeq2 does not share raw data, intermediate statistics may still pose privacy concerns. Additional measures like differential privacy or secure aggregation could be applied to further enhance security. Furthermore, standardized preprocessing protocols are essential for accurate comparisons across sites.
Researchers can explore its features and applications through the bioRxiv preprint and source code on GitHub.
Topics: Tools & Methods